Detailed Description
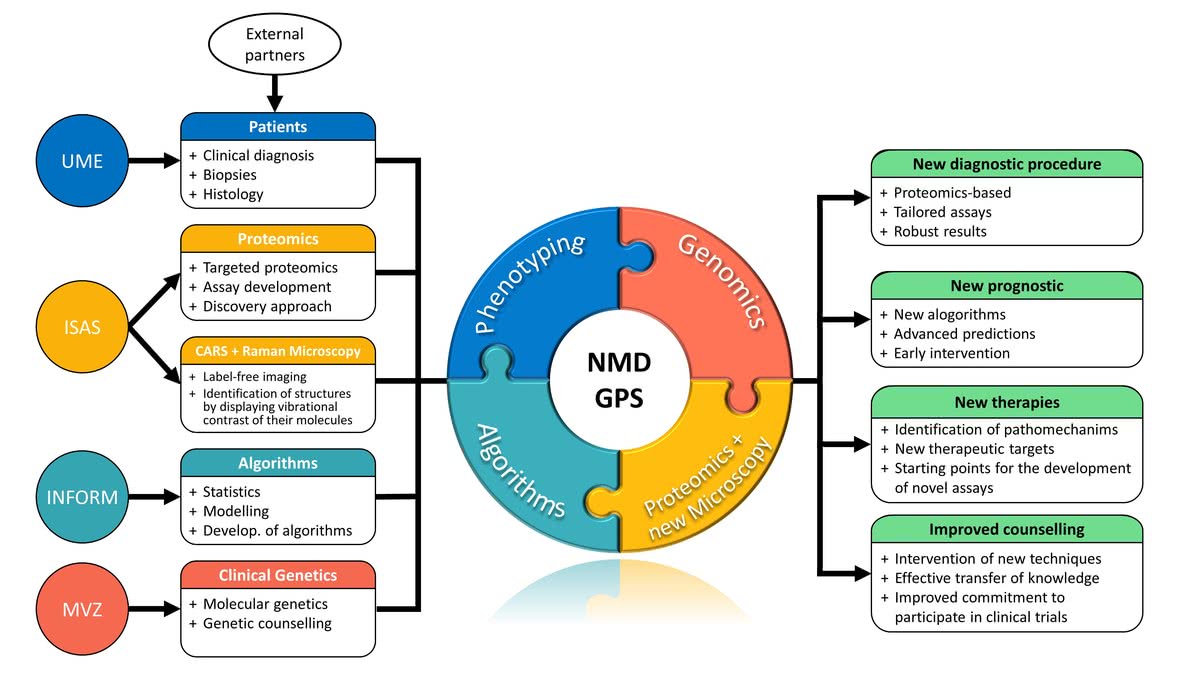
Generally, by addressing the need to
(i) replace the antibody-based immunological diagnostic analysis of muscle biopsy specimen derived from patients suffering from neuromuscular diseases (NMDs) as well as to
(ii) identify pathomechanisms common in different NMDs toward the definition of unified treatment concepts, NMD-GPS combines phenomics, genomics, proteomics and machine learning in an interdisciplinary manner.
For that purpose, precise clinical data derived from patients with a clinically and molecular genetic verified NMDs will be collected and a targeted proteomic profiling (quantifying proteins encoded by genes responsible for the clinical manifestation of a NMD or involved in the manifestation of a myositis) utilizing patient-derived muscle will be carried out. Hereby, stable-isotope labelled peptides will be used to warrant an absolute quantification which is more robust and precise compared to the antibody-based immunoblot analysis applied thus far. Results of this targeted measurement along with the intersection of molecular genetic data will allow to directly evaluate the effect of genetic mutations on protein abundances also by identifying patterns of co-regulation of proteins functionally connected or acting in concert. Notably, this approach is also very valuable to evaluate pathogenicity of ambiguous mutations frequently identified in the era of next generation sequencing. Consequently, in future, results of targeted proteomic profiling (in the framework of NMD-GPS) will also enable to predict the gene or inflammatory cascade causative for the phenotype of the individual patient. Hereby, the replacement of antibodies and immunoblot studies by stable-isotope labelled peptides and mass spectrometry based quantification of peptides (protein fragments) not only enables to address also proteins for which no suitable (or specific) antibody exists thus far but also significantly reduces the amount/ tissue required for this comprehensive diagnostic protein study. Additionally, unbiased (global) proteomic studies along with phosphoproteomic studies and the intersection of both by utilizing same muscle protein extracts as for the diagnostic study of NMD-causative proteins will be carried out. These studies will allow to identify pathways specific and common for the different entities caused by mutations in more than 300 genes linked to NMDs up to date. Here, intersection of exome sequencing data is crucial to exclude pathophysiological consequences based on the presence of mutations in further genes and to potentially identify modifying factors influencing the manifestation, progression or general severity of a NMD presenting with phenotypical variation. Results of these studies are crucial to define unified starting points for therapeutic intervention concepts, an important aspect to ambitionate pharma industry to develop drugs which can be used for a variety of NMDs grouped according to the underlying pathophysiology and overcome a significant problem in rare diseases and the research of same ones.
CARS microscopic studies utilizing muscle biopsy sections will allow to investigate general lipid and protein proportions in the tissue and hereby to identify accumulations and dysregulations/ dysproportions of both classes of biomolecules as spectral fingerprints of different NMDs. Thus, this approach represents a new concept in the microscopic-based classification of NMDs in the context of the diagnostic work-up of muscle biopsies by providing information beyond histology and enzyme activities.